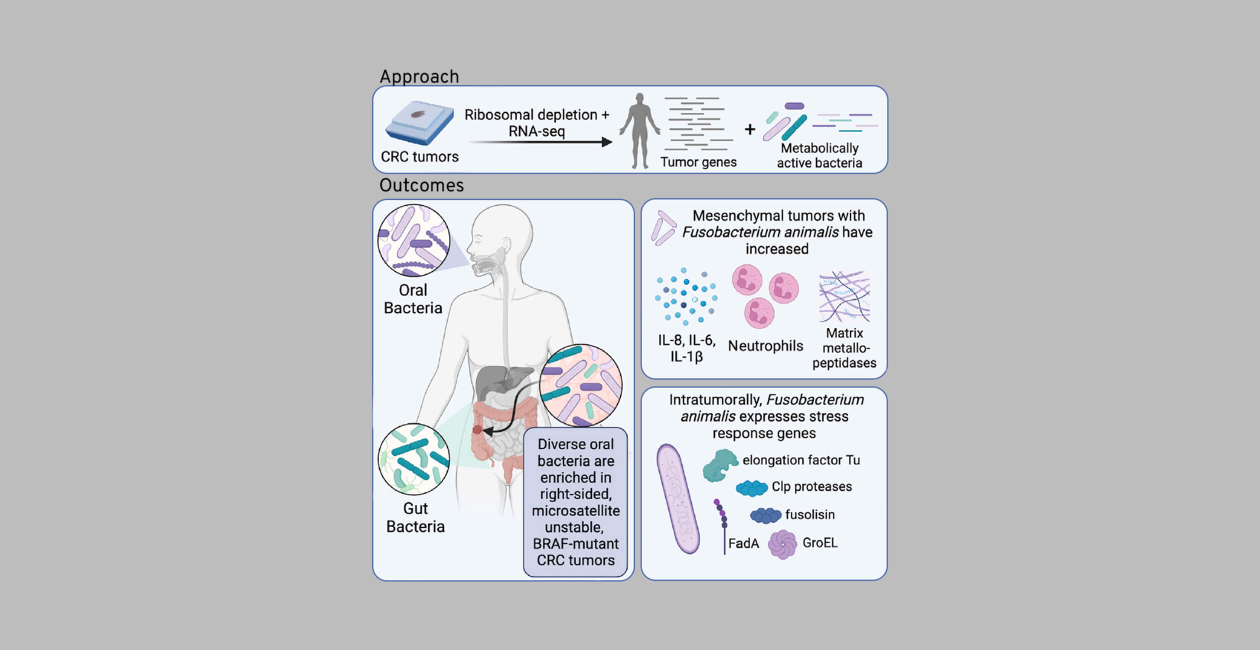
As well as studying the systemic effects of gut microbes as measured via their metabolites, the Byrd lab maintains an interest in understanding the local effects of bacteria to disease progression, therapeutic response, and immune system activation. To study this in colorectal cancer (CRC), where previous studies had used low resolution 16S ribosomal RNA sequencing or low depth microbial RNA-sequencing approaches, we leveraged an alternate approach to achieve deep coverage of microbial constituents of human tumors. Specifically, from the AVANT phase III trial, 807 CRC tumor samples were subjected to ribosomal depletion and metatranscriptomic RNA-sequencing in order to dually characterize tumor gene expression and intratumoral bacteria (Younginger et al. Cell Reports Medicine 2023). After extensive quality control to eliminate potential contaminants, we detected 74 diverse bacterial species. While the majority of these species were known gut commensals, 17 of them, including 4 Fusobacterium spp., were derived from the oral cavity and identified as enriched among right-sided, microsatellite instability-high (MSI-H), and BRAF-mutant tumors. Across tumor subtypes, integration of Fusobacterium animalis presence and tumor gene expression revealed that F. animalis had variable effects depending on the tumor context.
Moving forward, we will expand on these results by applying similar approaches and cutting-edge technologies to additional tumor samples as well as biopsies from inflamed and non-inflamed sites along the gastrointestinal tract.
Additional Research Projects
Steady State Microbiome and Metabolome